Gervaise Henry
Scan contact card
Skills
- AWS Certified Solutions Architect - Associate
- AWS Certified Cloud Practitioner
- Scrum Alliance Certified Scrum Product Owner
- R
- Python
- Nextflow
- WDL
- git
- linux
- bash
- Static Site Generators
- Serverless Deployment
- GitLab Pages
- html5
- css3
- javascript
- amazonwebservices
- Azure
- googlecloud
- S3
- EC2
- Batch
- Lambda
- DynamoDB
- API Gateway
- Certificate Manager
- Route53
- CloudFront
- CloudTrail
- Athena
- Docker
- Singularity
- bulkRNA-Seq
- Gene Expression Microarray
- scRNA-Seq
- scTCR-seq
- Spatial Transcriptomics
- Variant Calling
- Proteomics (Mass Spec)
- Varied Molecular Biology Techniques
GUDMAP/RBK Analytics

Design and manage the Agile group responsible for creating analytics for data stored on the NIH consortiums GUDMAP and RBK. This analytics are not only best-practices pipelines written in Nextflow, but also integrates seamlessly with the consortiums' data-hub, in terms of initiation, and data ingress/egress. It is built with the flexibility to utilize on premises HPC, or a custom built AWS architecture (including many serverless resources) for low-cost, highly-available queueing, compute, and reporting. It can also be easily adapted to almost any running environment using custom configs. The entire pipeline utilizes dockerized micro-services which is orchestrated by Nextflow.
Nextflow in the Cloud
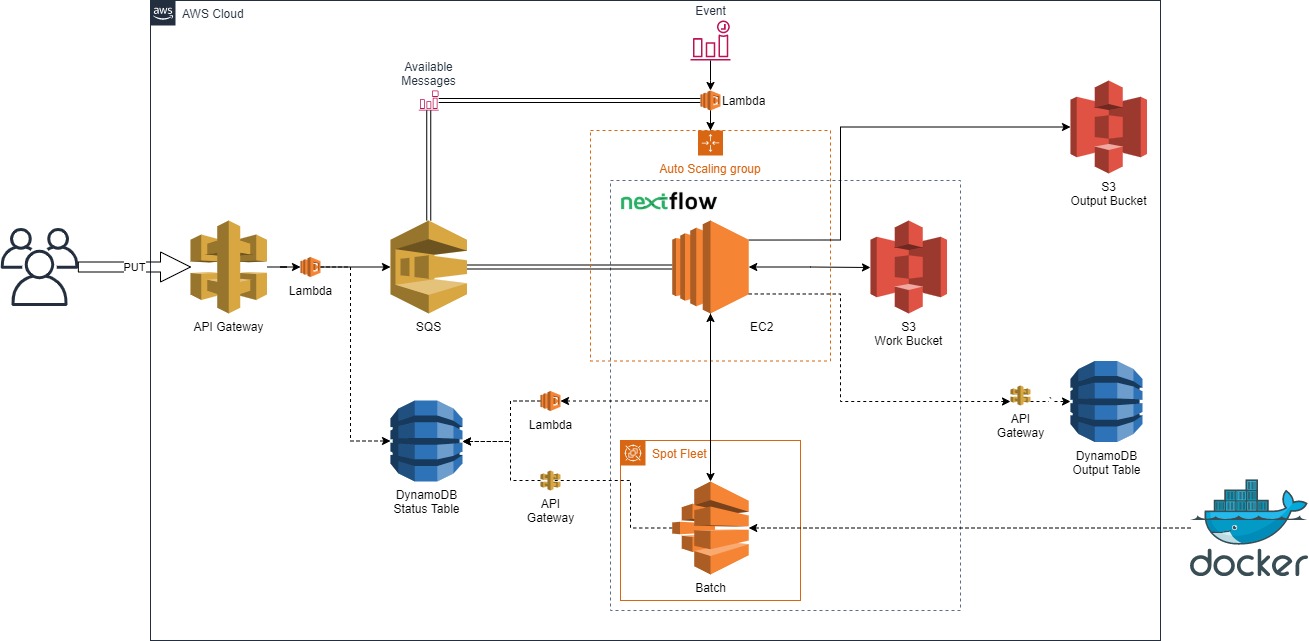
Manage a small agile team to create a proof of concept AWS architecture to run Nextflow pipelines in the cloud. It utilizes low-cost, highly-availability queueing, compute, and storage resources. (NOTE: project decomissioned and the git repository is no longer public, for more information feel free to reach out to [Gervaise Henry](ghenry@gmail.com))
Pipeline Run Tracking
Creating a pipeline run tracking tool for the Bioinformatics Core Facility at UT Southwestern Medical Center. This tool utilizes AWS serverless offerings, including REST-API and a website for run tracking. (NOTE: project decomissioned and the git repository is no longer public, for more information feel free to reach out to [Gervaise Henry](ghenry@gmail.com))
Strand Lab External Website
The Strand Lab website was designed to not only provide a location to share information about the lab, but also provide a means to showcase the lab's data and biorepository. The single-cell data is displayed using CZI's cellxgene visualization tool as well as pre-generated genome-wide visualization of the expression data. The lab has an extensive biorepository of human lower urinary tract biosamples and de-identified clinical data as well as H&E images are available for viewing and filtering on the website. The site is hosted serverlessly on AWS.
Experience
VP Data Engineering
Leads data infrastructure and management strategy. Responsible for designing, implementing, and maintaining a robust data platform that supports Champions’ research and development efforts, with a strong focus on managing multi-omics and research data in compliance with FAIR principles.
Head, Professional Services Engineering
- Manage a large organization of diverse technical global staff:
- Across the US, Europe, and Asia.
- Solutions Architects.
- Services Bioinformatics Engineers.
- Services Contractors.
- Team responsible for all tool and workflow development as a service for customers.
- Team responsible for ingenstion for translational datasets as a service for customers.
- Team responsible for building external integrations, including customer cloud integrations, and software development, to automate and layer novel funtionality to the DNAnexus platofrm.
- Responsible for staffing projects, including the use of a large contigent of off-shore contractors (not limited to those directly reporting to the PS Engineering Organization).
- Work with other stakeholders in developing process to ensure profitable services.
- Work closely with Product Engineering and Product Management teams to assist in scoping and prioritizing new features and functionality.
- Build proof-of-concept assets to assist in architecting new features and functionality.
- Assist in technical evaluation of potential partners, as well as building proof-of-concept integrations when necessary.
- Act as a liaison between the sales team, the customer, and internal product, and support teams.
Manager, Solutions and Services Engineering
- Manage 13 people (across the US and Europe) including:
- 2 group managers.
- 2 solutions architects.
- 3 solutions engineers.
- 9 services engineers.
- Responsible for all tool and workflow development, in both pre-sales and professional services settings.
- Responsible for all ingenstion for translational datasets, in both pre-sales and professional services settings.
- Responsible for staffing projects, including the use of a large contigent of off-shore contractors.
- Work with other stakeholders in developing process to ensure profitable services.
- Work closely with Product Engineering and Product Management teams to assist in scoping and prioritizing new features and functionality.
- Build proof-of-concept assets to assist in architecting new features and functionality.
- Assist in technical evaluation of potential partners, as well as building proof-of-concept integrations when necessary.
- Act as a liaison between the sales team, the customer, and internal product, professional services, and support teams.
Manager, Solutions Engineer
- Manager of presales engineering group:
- Manage a team if 5-6 Solutions Engineers.
- Build proof-of-concept assets to prove the customer’s need are possible on the platform, both scientifically and technically.
- Assist in scoping a customer’s scientific and technical needs.
- Perform platform demos to customer.
- Provide technical customer training in a presales setting.
- Provide technical support in a presales setting.
- Work closely with Product Engineering and Product Management teams to assist in scoping and prioritizing new features and functionality.
- Build proof-of-concept assets to assist in architecting new features and functionality.
- Act as a liaison between the sales team, the customer, and internal product, professional services, and support teams.
Senior Solutions Engineer
- Presales engineer:
- Assist with customer discovery.
- Scope and scale customer needs and wants to adiquately demmonstrate platform functionality.
- Build demo, proof-of-concept, and pilot assets.
- Perform demontrations of the platform to customers.
- Provide customer training.
- Provide white-glove technical support experience for potential strategic accounts.
- Act as liaison between sales team, customer, and external partners with internal product and services teams.
Computational Biologist
- Big-Data Analysis: Responsible for producing high quality statistical analysis for a wide range of data types, in particular, genomics-based modalities. This is dominated by the analysis of large amounts of singe-cell RNA-sequencing data.
- Tool Development: Created tools (including shiny-apps) for data exploration by the lab.
- Website Development: Create and manage the lab website using static site generators. The website includes custom tools for exploring the lab’s biorepository and scRNA-seq data, and serverless deployment on AWS, as well as gitlab pages.
- Pipeline Development: In collaboration with the Bioinformatics Core Facility (BICF), I help to create and manage automated pipelines to ensure reproducibility, transparent analysis, and to increase analytical throughput.
- Collaboration Management: Manage a funded collaboration to create pipelines for standardizing data analysis for the intent of integration of data from disparate studies on a consortium level. This includes designing the pipelines, contributing to the development, and managing a team of developers.
- Cloud Architect: Migrated a variety of BICF pipelines to function seamlessly in the cloud (including AWS and Azure).
- Instructor: Contributed regularly in the instruction of the Department of Bioinformatics computational nano-courses across multiple topics including programing languages and analytic techniques.
- RESULTS: Published three pipelines, with an additional four in active development. Secured consortium level funding for pipeline development. Successfully migrated 2 pipelines and the Strand Lab website to AWS, and actively migrating remaining pipelines.
Research Associate
- Research: Designed, conducted, and analyzed cellular and molecular experiments, leveraging best practices, statistical techniques, and available resources to maximize outcomes and publish impactfully.
- Author: I presented annually at conferences, both in poster and oral formats. I was a named author in nine journal publications, three of which I was the primary researcher and the first author.
- RESULTS: Authored (first, and co-authored) 8 journal papers, including a very successful first-author Cell Reports (2018) paper.
Research Associate
- Assay Development: Develop high-content image-based assays for high throughput drug discovery.
- RESULTS: Optimized high throughput assays for early-stage drug discovery on marine natural products.
Teaching Assistant/Research Assistant
- Research: Utilized cellular and molecular techniques to further projects.
- Supervision: Supervised several of undergraduate researchers to sustain collaborations.
- Instructor: Independently taught freshman and senior level lab courses. Responsible for assisting with design of laboratory content including assessments, independently teaching lab content, grading, and counseling students.
- RESULTS: Successfully defended master’s thesis linking obesity and BMI to the progression of multiple myeloma. Contributed to the success of three lab projects not associated with master’s work.
Research Technician
- Research: Conducted research to support several projects.
- Vendor Management: Managed the lab budget, oversaw ordering, and developed vendor relationships.
- RESULTS: Successfully allowed for the seamless transition of the new Pathology Department chairman’s lab to UT Southwestern.
Screening Scientist
- High-Throughput Screening: Conducted high throughput compound and whole genome RNAi screens for client labs.
- Assay Development: Involved in all aspects of projects including assay choice and development, optimization, implementation, and data analysis.
- RESULTS: Contributed to successful screens, which led to significant lead-generation and fueled many research projects.
cellxgene: Single cell analysis of mouse and human prostate reveals novel fibroblasts with specialized distribution and microenvironment interactions

Single-cell RNA-sequencing of adult human prostates and urethras from organ donors, and BPH (glandular and stromal) patients as well as adult mouse lower urinary tracts.
RNA-seq Analytic Pipeline for GUDMAP/RBK

This pipeline conducts end-to-end RNA-seq analysis on replicates stored in the GenitoUrinary Development Molecular Anatomy Project and ReBuilding the Kidney consortium. This pipeline was created in a collaboration between the Strand Lab and Bioinformatics Core Facility (BICF) at UT Southwestern Medical Center.This pipeline conducts end-to-end RNA-seq analysis on replicates stored in the GenitoUrinary Development Molecular Anatomy Project and ReBuilding the Kidney consortium. This pipeline was created in a collaboration between the Strand Lab and Bioinformatics Core Facility (BICF) at UT Southwestern Medical Center.
Strand Lab analysis of single-cell RNA sequencing

This code was used for the single-cell RNA-sequencing analysis in the Strand Lab at UT Southwestern Medical Center
BICF Cellranger count Analysis Workflow

BICF Cellranger count Analysis Workflow is a wrapper for the cellranger count tool from 10x Genomics. This pipeline, used by the Bioinformatics Core Facility at UT Southwestern, takes fastq files from 10x Genomics single-cell gene expression libraries and passes them to cellranger count, managing parallelization of multiple runs, as well as, aggregation as appropriate. This pipeline is primarily used with a SLURM cluster on BioHPC, but it should be able to run on any system that Nextflow supports. Additionally, this pipeline is designed to work using a simple web interface.
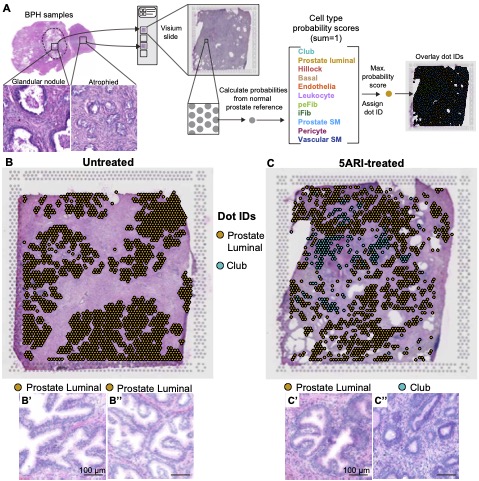
GEO: Human prostate luminal epithelia adopt a club-like state in response to low androgen levels

Spatial transcriptomics of treatment naive and 5-alpha reductase inhibitior treated human BPH tissue
GEO: Single cell RNA-sequencing of verumontanum region and peripheral zone of normal human prostate

Dissected verumontanum region and peripheral zone region from a normal human prostate sample were digested into single-cell suspensions, cells were subject to single-cell RNA-sequencing using the 10x Genomics platform
GEO: Single-cell RNA-sequencing of adult human normal and BPH (glandular and stroma) prostates and urethra

Single-cell RNA-sequencing of adult human prostates and urethras from organ donors, and BPH (glandular and stromal) patients.
GEO: Single-cell RNA-sequencing of adult mouse lower urinary tracts v2

GUDMAP: Single cell analysis of mouse and human prostate reveals novel fibroblasts with specialized distribution and microenvironment interactions

Sequencing data relating to the in preparation publication: 'Single cell analysis of mouse and human prostate reveals novel fibroblasts with specialized distribution and microenvironment interactions'
The Journal of Pathology - 5-alpha reductase inhibitors induce a prostate luminal to club cell transition in human benign prostatic hyperplasia
Joseph DB, Henry GH, Malewska A, Reese JC, Mauck RJ, Gahan JC, Hutchinson RC, Mohler JL, Roehrborn CG, Strand DW
The Journal of Pathology - Single cell analysis of mouse and human prostate reveals novel fibroblasts with specialized distribution and microenvironment interactions
Joseph DB, Henry GH, Malewska A, Reese JC, Mauck RJ, Gahan JC, Hutchinson RC, Malladi VS, Roehrborn CG, Vezina CM, Strand DW
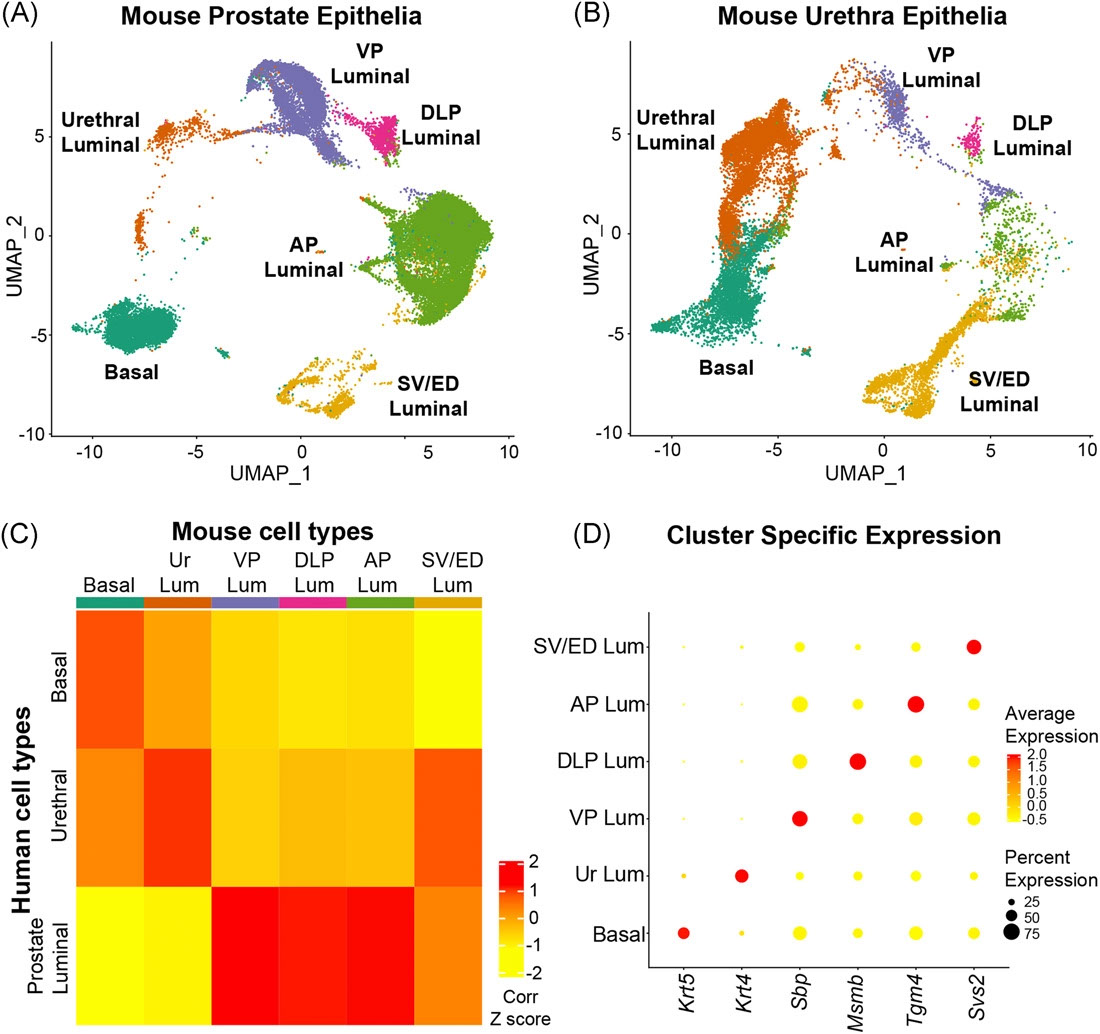
The Prostate - Urethral luminal epithelia are castration-insensitive cells of the proximal prostate
Joseph DB, Henry GH, Malewska A, Iqbal NS, Ruetten HM, Turco AE, Abler LL, Sandhu SK, Cadena MK, Malladi VS, Reese JC, Mauck RJ, Gahan JC, Hutchinson RC, Roehrborn CG, Baker LA, Vezina CM, Strand DW
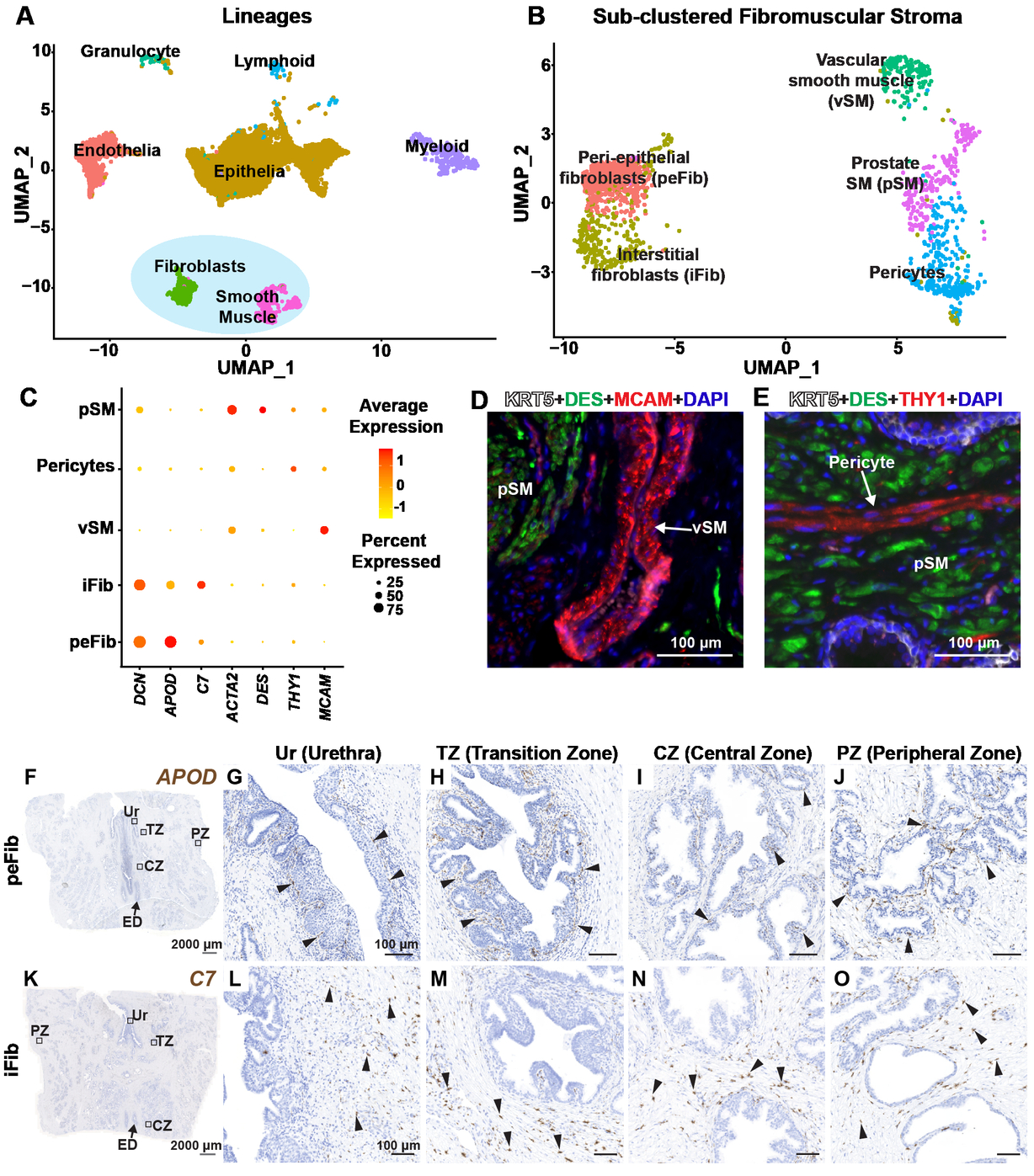
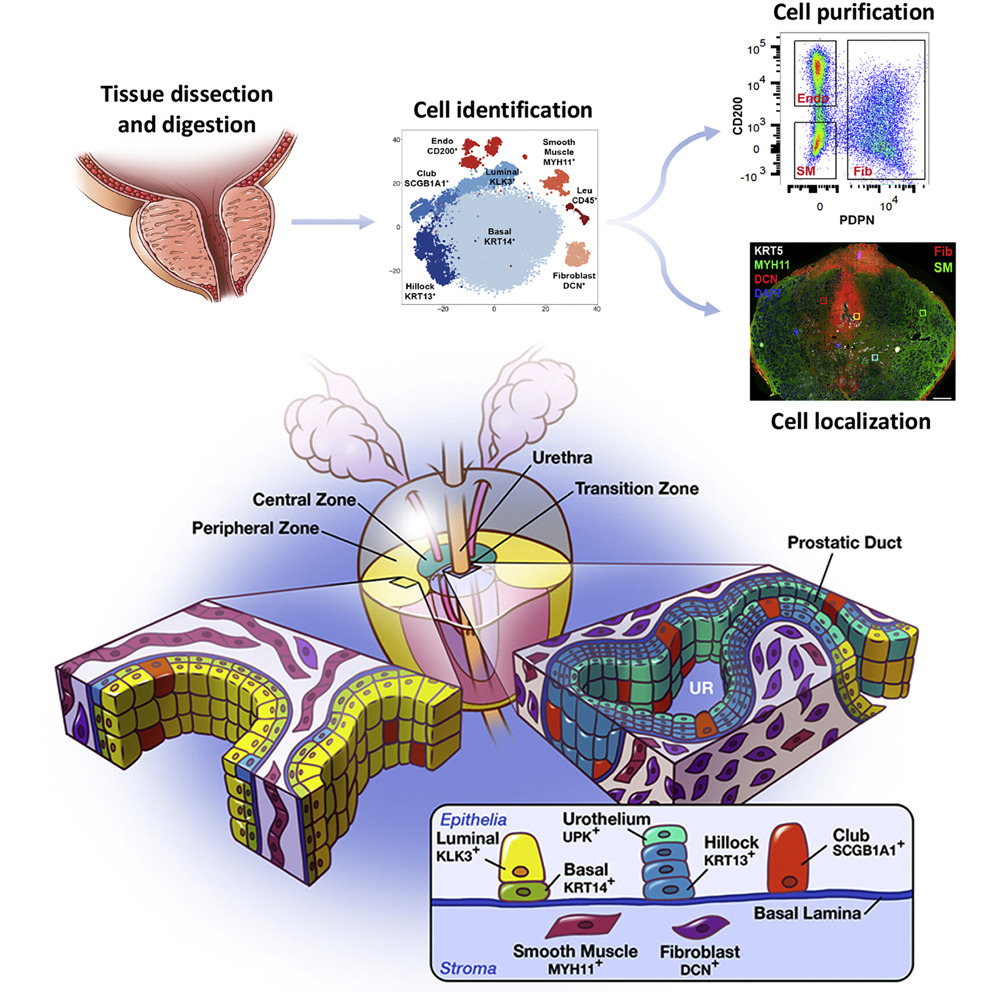
Cell Reports - A cellular anatomy of the normal human prostate
Henry GH, Malewska A, Joseph DB, Malladi VS, Lee J, Torrealba J, Mauck RJ, Gahan JC, Raj GV, Roehrborn CG, Hon GC, MacConmara MP, Reese JC, Hutchinson RC, Vezina CM, Strand DW
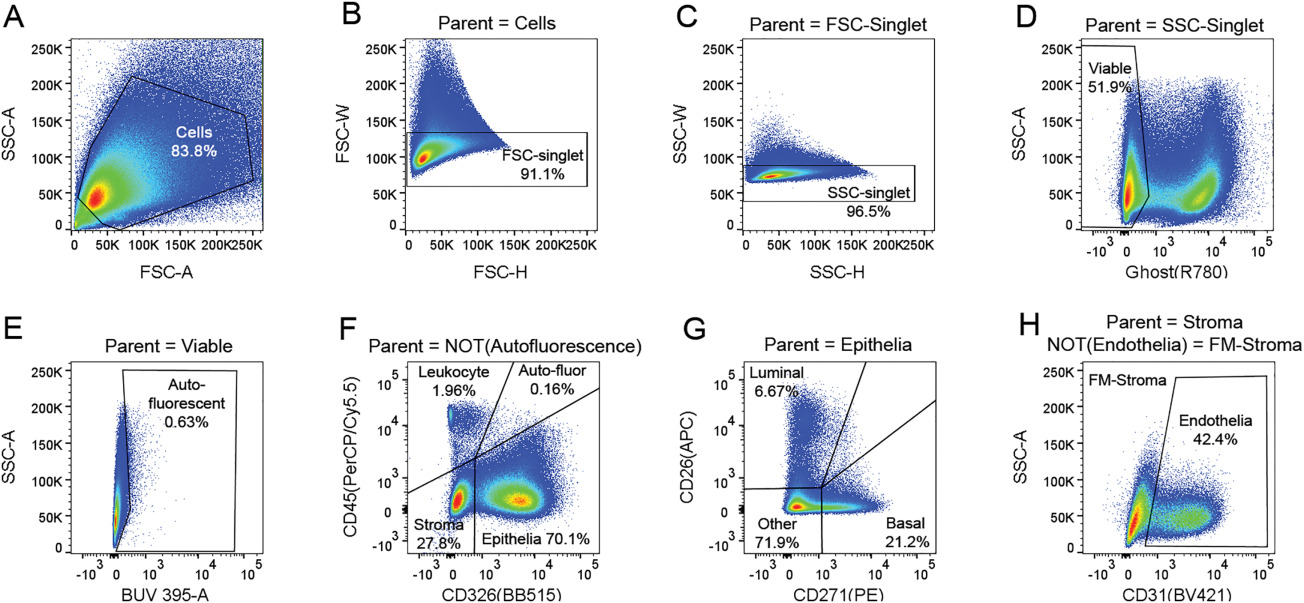
Cytometry Part A - OMIP-040: Optimized gating of human prostate cellular subpopulations
Henry GH, Loof N, Strand DW
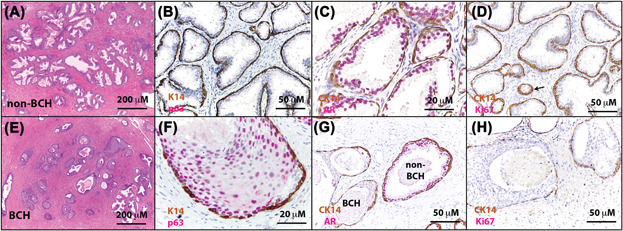
The Prostate - Molecular pathogenesis of human prostate basal cell hyperplasia
Henry GH, Malewska A, Hutchinson R, Gahan J, Mauck R, Francis F, Torrealba J, Roehrborn C, Strand DW
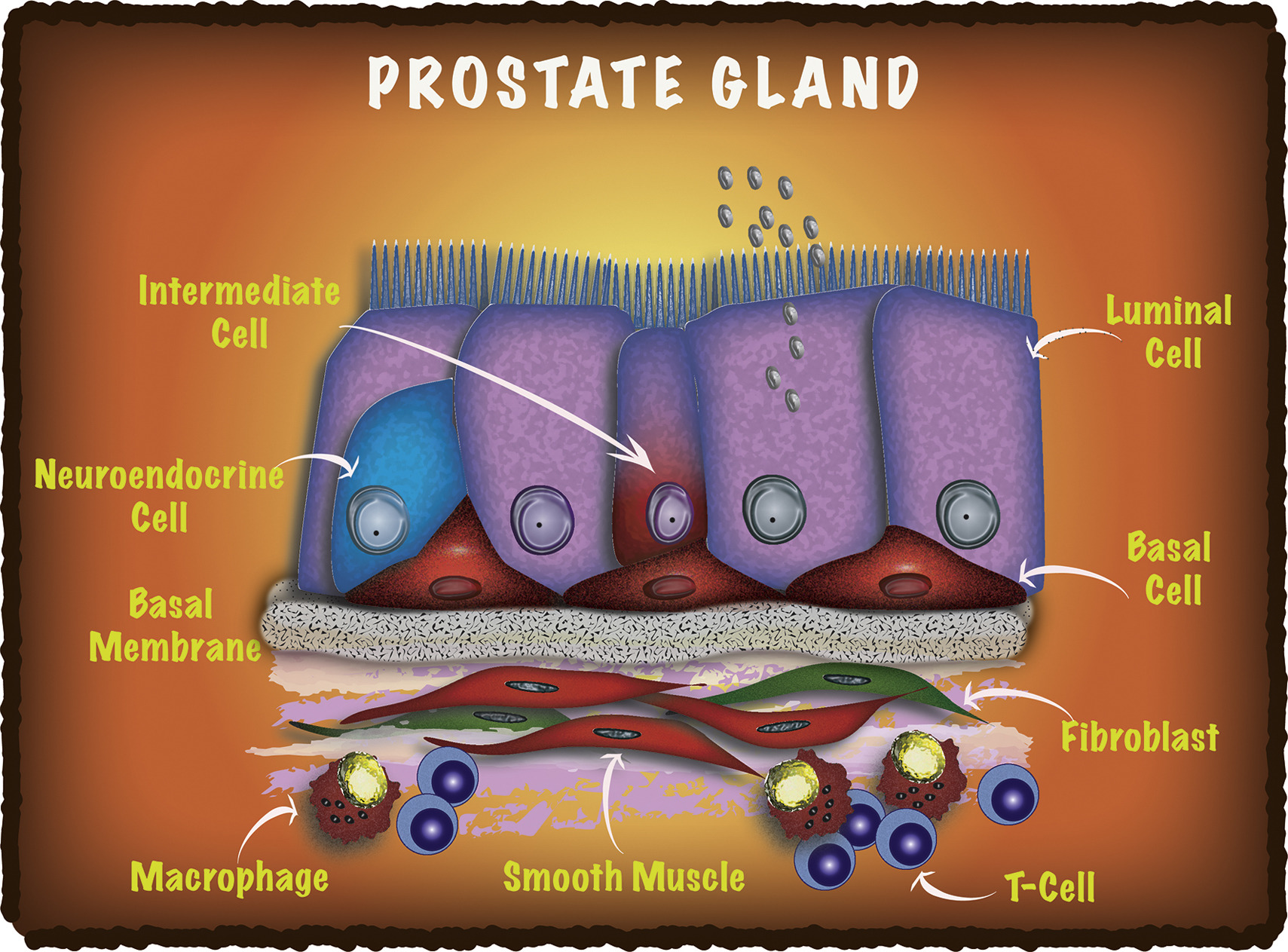
Differentiation - Isolation and analysis of discrete human prostate cellular populations
Strand D W, Aaron L, Henry G, Franco O E, Hayward SW
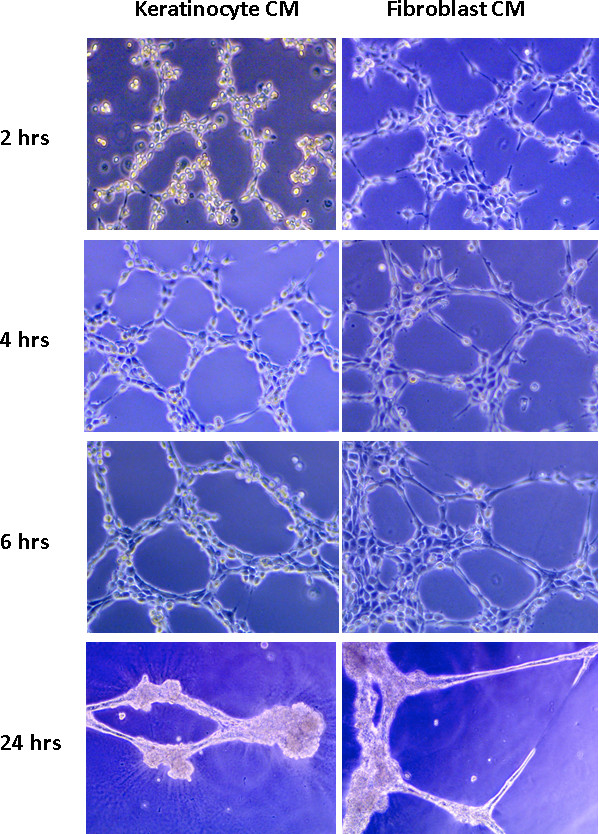
JoVE - Endothelial cell tube formation assay for the in vitro study of angiogenesis
DeCicco-Skinner KL, Henry GH, Cataisson C, Tabib T, Gwilliam JC, Watson NJ, Bullwinkle EM, Falkenburg L, O’Neill RC, Morin A, Wiest JS
EU Biobank Week 2024 - April 2024
Efficiently Analyzing Population-Scale Genomics Data
Sedlakova, A, Henry GH
ACMG - March 2024
Automated Genome Interpretation Solution to Accelerate Diagnostic Decisions via Intelliseq GeneSpect™ Reporter
Szklarczyk-Smolana K, Pacewicz K, Henry GH
BioTechX Europe - October 2023
Bridging the Gap: Solutions for Data Integration, Security, and Collaborative Exploration in Precision Medicine
Rao A, Henry GH
Precision Medicine Leaders Summit: Advances in Single Cell & Spatial Biology - September 2023
Democratizing Access to Single-Cell RNA Sequencing with the Parse Biosciences Evercode Platform
Henry GH, Roco C
Precision Medicine Leaders Summit: Advances in Single Cell & Spatial Biology - September 2023
Innovation Panel: Building the Spatial Biology Ecosystem
Enderlein C, Clark JN, Emanuel G, Henry GH, Kim J
BioIT World - May 2023
Towards Precision Medicine: Datasets, Computation, and Data Integration for Patient Subsetting Research - Part I
Busby BR, Chu E, Floden E, Henry GH, Huitt E
BioIT World - May 2023
Achieving FAIRness with Multimodal Data
Busby BR, Dawson E, Gillis M, Henry GH, Holt R, Neilley V
Cold Spring Harbor: Single Cell Analysis - November 2019
A Cellular Atlas of Human Prostate Disease
Henry GH, Malewska A, Malladi VS, Lee J, Gahan JC, Reese J, Strand DW
American Urological Association: Annual Meeting - May 2019
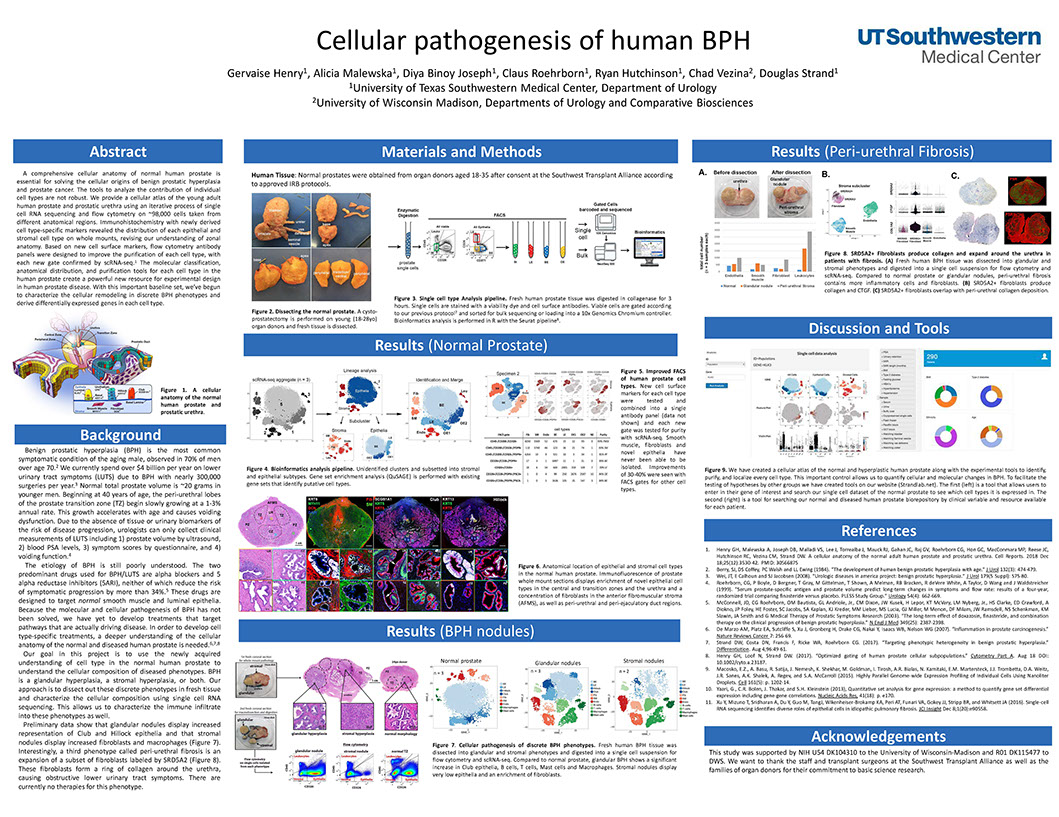
Cellular pathogenesis of human BPH
Henry G, Malewska A, Binoy Joseph D, Roehrborn C, Hutchinson R, Vezina C, Strand D
Collaborating for the Advancement of Interdisciplinary Research in Benign Urology: Meeting - December 2018
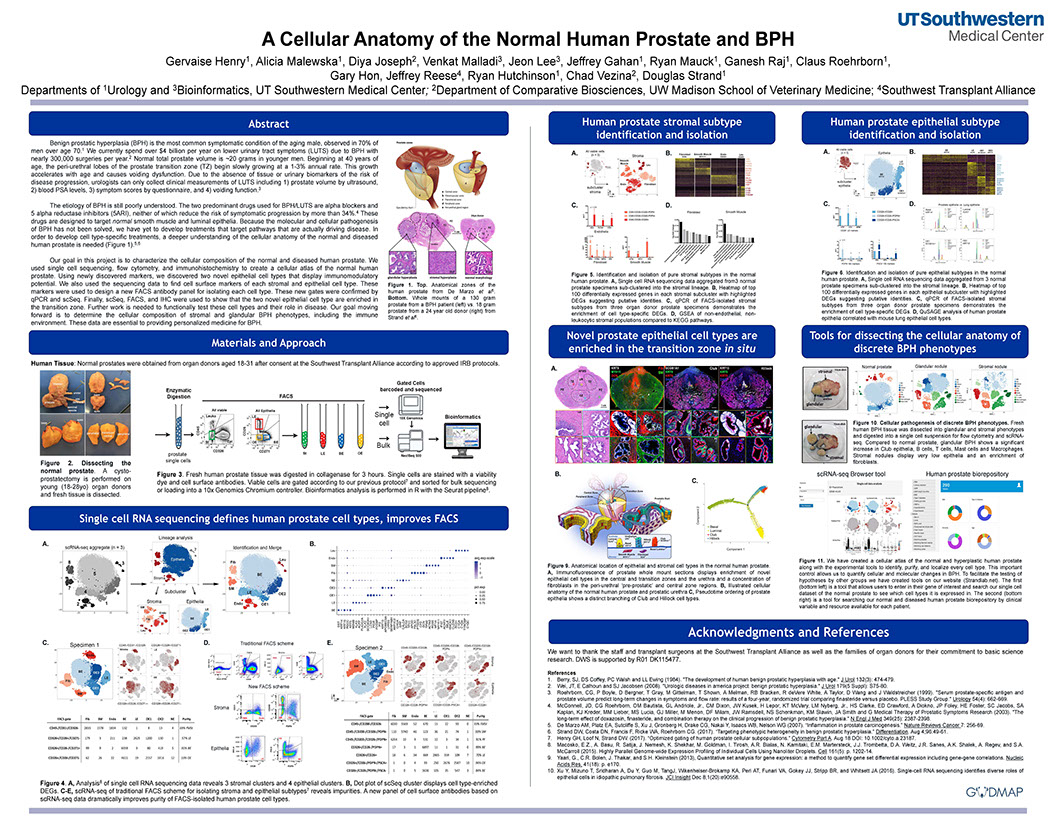
Building a comprehensive cellular anatomy of the normal and diseased human prostateia
Henry GH, Malewska A, Joseph DB, Malladi VS, Lee J, Torrealba J, Mauck RJ, Gahan JC, Raj JV, Roehrborn CG, Hon GC, MacConmara MP, Reese JC, Hutchinson RC, Vezina CM, Strand DW
Society for Basic Urologic Research: Annual Meeting - November 2018
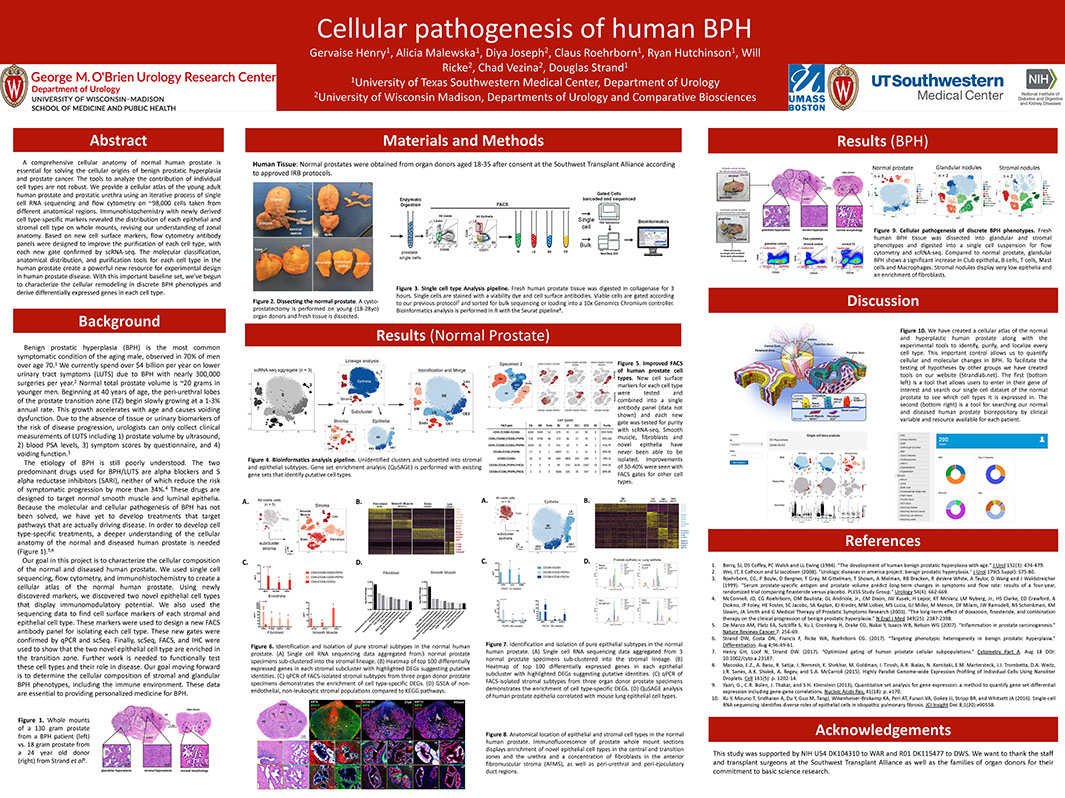
A Cellular Anatomy of the Normal Human Prostate and BPH
Henry G, Malewska A, Binoy Joseph D, Malladi V, Lee J, Gahan J, Mauck R, Raj G, Roehrborn C, Hon G, Reese J, Hutchinson R, Vezina C, Strand D
10x Genomics User Group Meeting: Houston - May 2018
Determining Cellular Heterogeneity in Human Prostate using the Chromium™ Single Cell 3’ Solution
Henry GH
FlowTex Conference - February 2018
Determining cellular heterogeneity in human prostate with flow cytometry and single-cell RNA-sequencing
Henry GH
North Texas Flow Cytometry Conference - December 2017
Determining cellular heterogeneity in human prostate with flow cytometry and single-cell RNA-sequencing
Henry GH
Society for Basic Urologic Research: Annual Meeting - November 2017
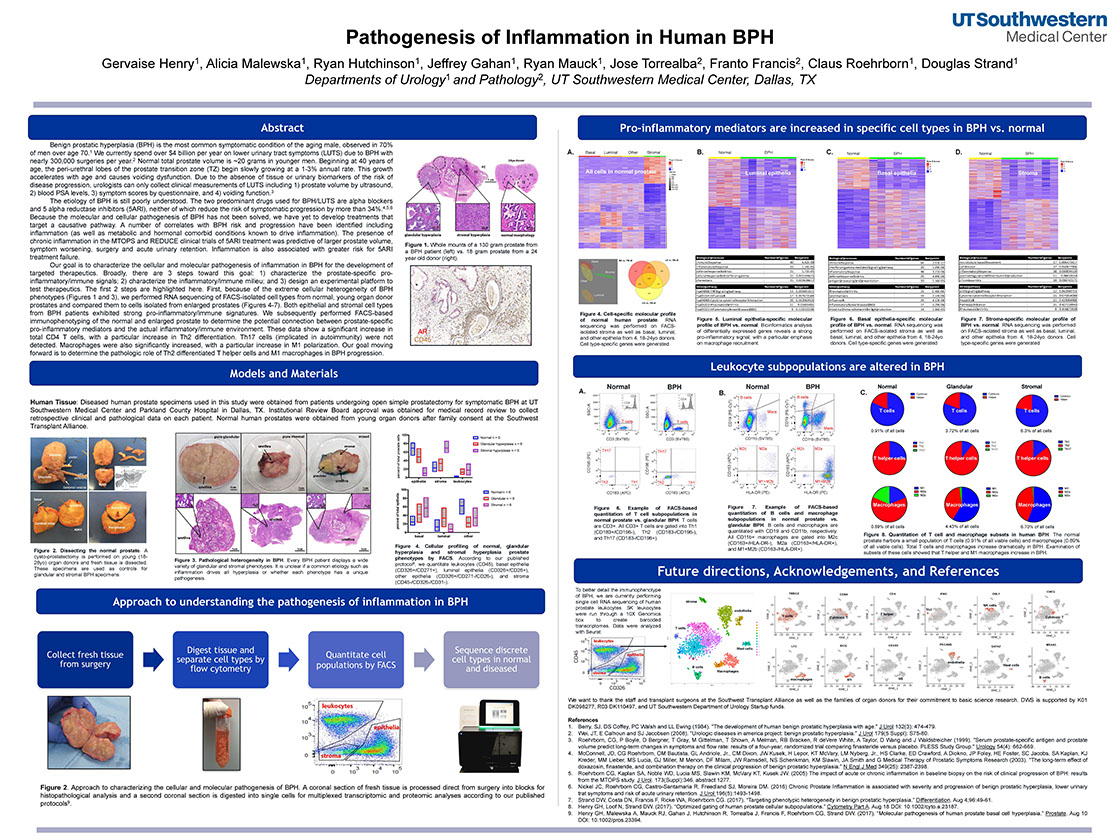
Pathogenesis of Inflammation in Human BPH
Henry G, Malewska A, Hutchinson R, Gahan J, Mauck R, Torrealba J, Francis F, Roehrborn C, Strand D
North Texas Flow Cytometry Conference - December 2016
Deconstructing BPH Phenotypes: Molecular Pathogenesis of Basal Hyperplasia
Henry GH
Society for Basic Urologic Research: Annual Meeting - November 2016
Deconstructing BPH phenotypes
Henry G, Malewska A, Mauck R, Torrealba J, Roehrborn C, Strand D
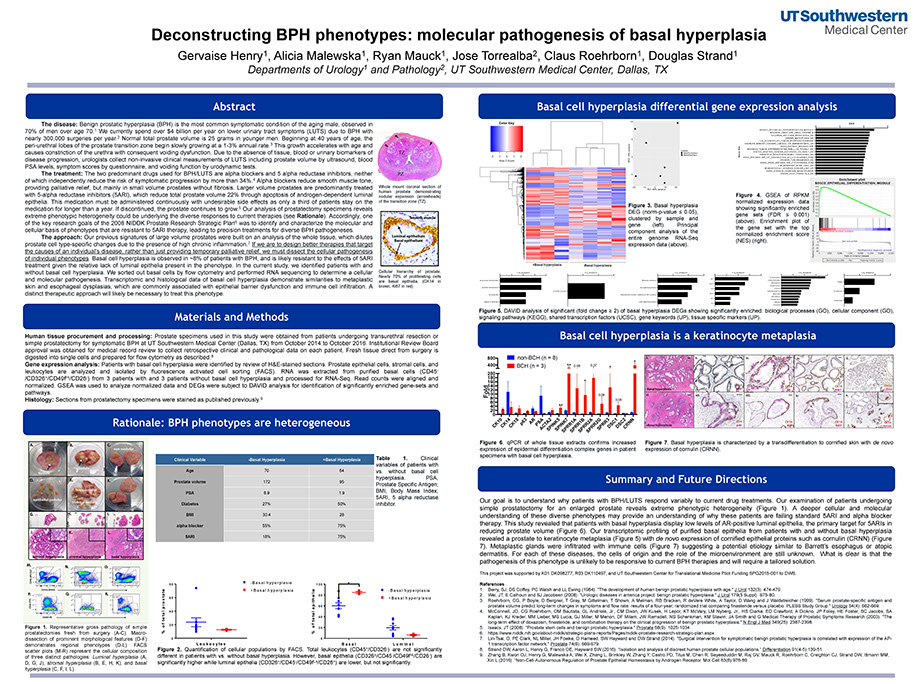
Society for Basic Urologic Research: Annual Meeting - November 2015
Cell-specific responses to inflammation in BPH
Henry G, Malewska A, Mauck R, Roerborn C, Strand D
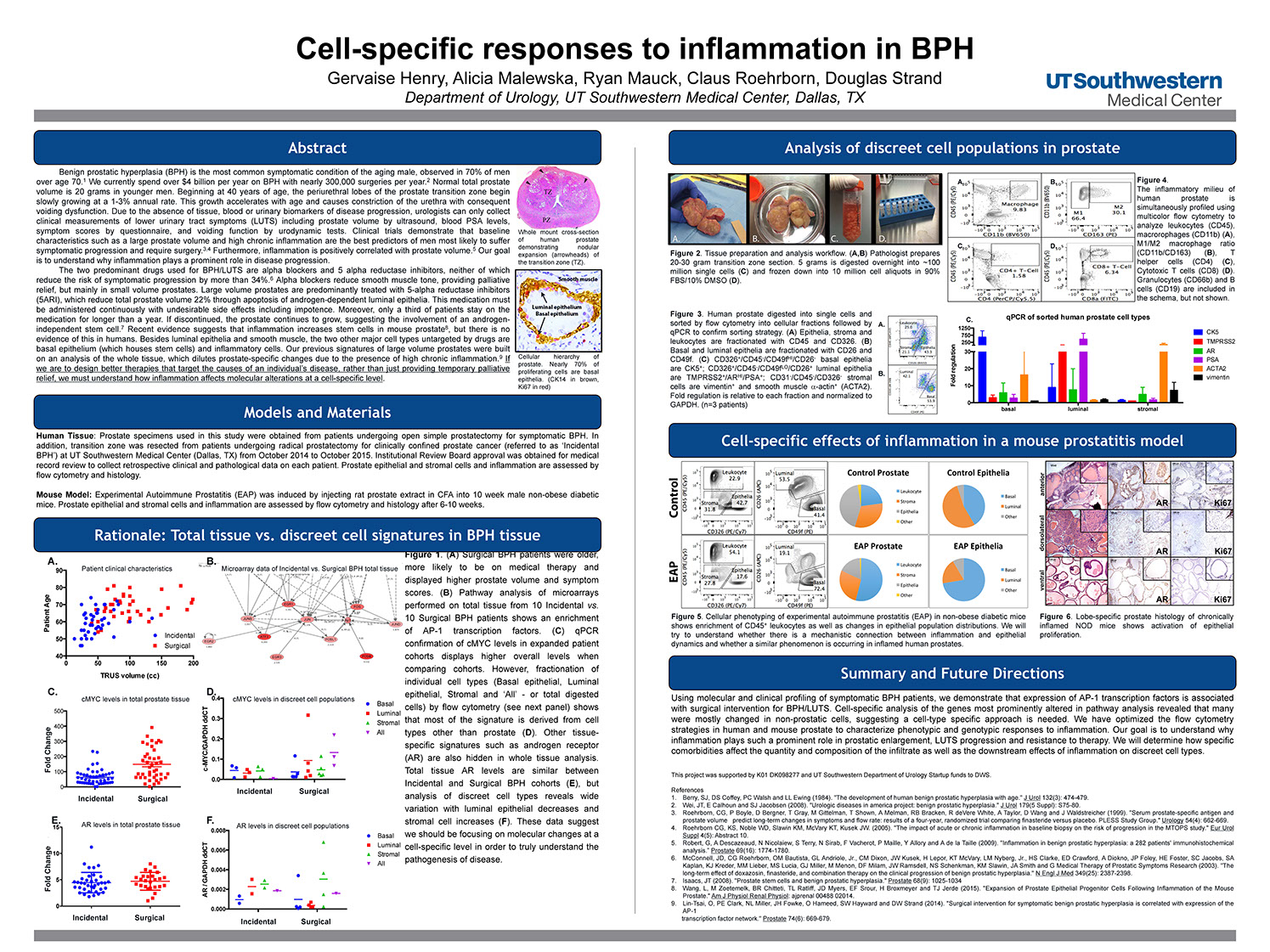
American Association of Cancer Research: Annual Meeting - April 2014
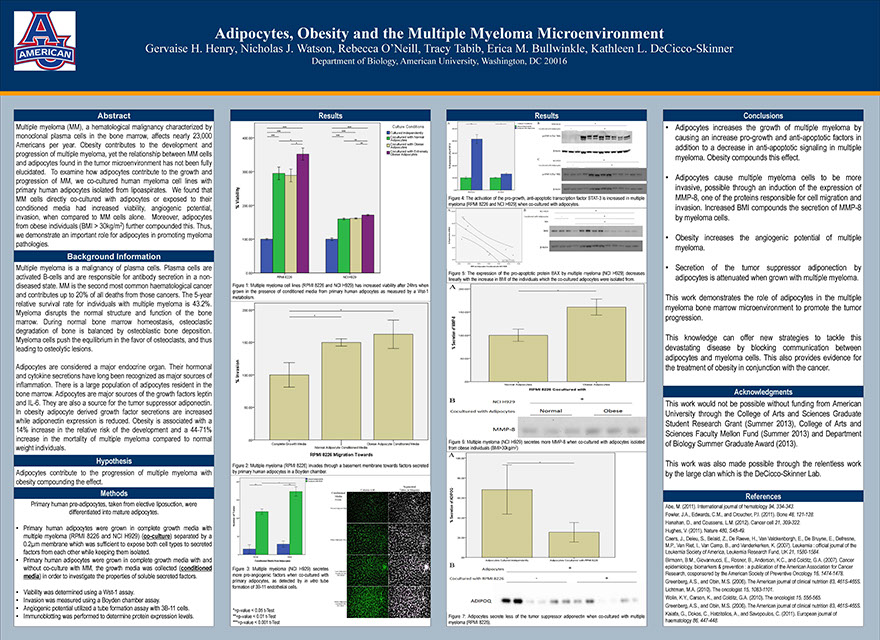
Adipocytes, Obesity and the Multiple Myeloma Microenvironment
Henry GH, Watson NJ, O’Neill R, Tabib T, Bullwinkle EM, DeCicco-Skinner KL